|
MOLECULAR DYNAMICS SIMULATIONS OF RHODOPSIN
AND PRION PROTEINS: THE EFFECT OF DISEASE-RELATED AMINO ACID
MUTATIONS ON THE STRUCTURAL CONFORMATIONS
Kholmurodov K. T.
Molecular Dynamics (MD) simulations were performed to investigate
the structural conformation of the rhodopsin and human prion proteins.
We have estimated the effect of specific disease-related
amino acid mutations on the dynamics and conformational changes.
For the rhodopsin (Rh) protein we performed 3-ns simulations and presented
the structure analysis data for the dark-adapted state of Rh. The RMSD (root-mean-square deviation) data for helices I-VII of Rh were evaluated.
From the analysis of the RMSDs we observed a relative movement of Rh helices,
which possesses not a similar behavior for all the helices.
Helices I, III and V exhibit the highest deviations from the reference structure.
The stucture images of Rh were constructed and the representative
configurations were compared for the 3-ns state. The results show the largest displacements from the reference structure for helices III and V.
The largest deviations are seen in the cytoplasmic region of these helices,
the extracellular ends show slight movement. We have also simulated the effect of the E134N mutation on the Rh conformation dynamics.
The E134N mutation has been considered to be important in the functionality of Rh.
We have evaluated the RMSD behavior for the wild-type Rh protein and Rh with E134N mutation. The comparison of the analysis data indicated that the reference and E134N mutant structures are subjected to a similar behavior. The relative helical movement of the Rh protein seems to be sensitive mostly in the cytoplasmic region.
The MD simulations were performed on three models of the human PrP (about 2 million time step/each structure) to investigate the effect of the specific
disease-related point mutations on the conformation dynamics.
We observed a partial rearrangement of the prion structure due to a
Glu200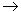 Lys mutation,
and provided a detailed mechanism of this process. Lys mutation,
and provided a detailed mechanism of this process.
PDF (1.38 Mb)
|
